Create genome browsers for the interactive annotation of any eukaryotic genomes
Web-based platform
G-OnRamp provides an integrated, web-based, graphical interface for performing large scale bioinformatics analyses.Designed for any genome
G-OnRamp is designed to work with any eukaryotic genomes.Reproducible workflow
G-OnRamp record all the steps in the analysis workflow (e.g., tool parameters, data conversions) so that you can apply the same workflow to other eukaryotic genomes.Share your results
The G-OnRamp analysis history (datasets, tools, and parameter settings) can be shared publicly or only with your colleagues.Customizable
You can incorporate additional Galaxy tools and modify the G-OnRamp workflow (e.g., tool parameters) according to your needs.Multiple deployment options
G-OnRamp analysis can be done on the cloud (e.g., Amazon EC2) or on the a local virtual machine (e.g., VirtualBox) instanceGEP + Galaxy = G-OnRamp
The G-OnRamp project combines two successful and long-running projects—the Genomics Education Partnership (GEP) and the Galaxy Project. It aims to create an integrated, web-based, and scalable environment that enables biologists to utilize large genomic datasets for the interactive annotation of any eukaryotic genome.
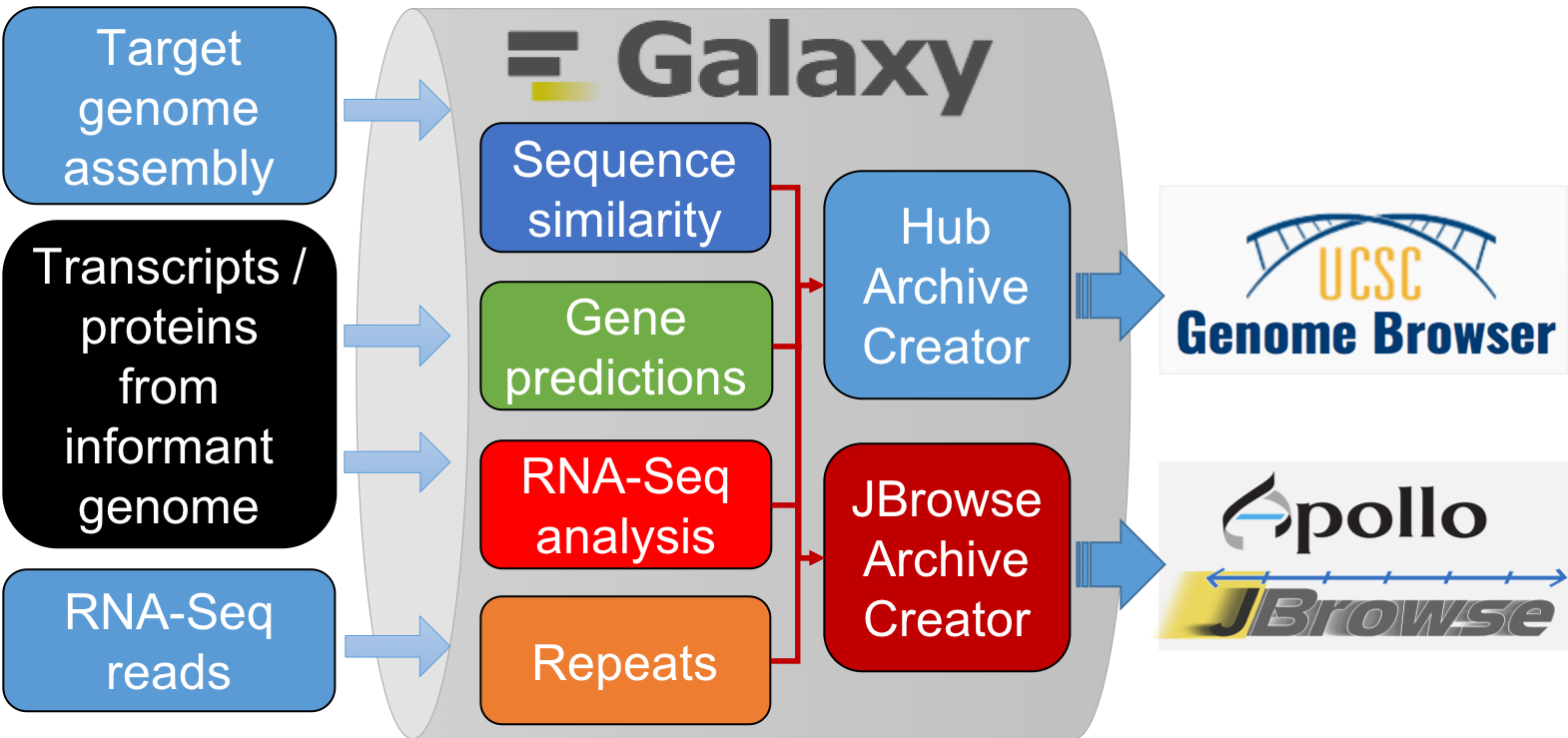
From genome data to genome browser:
The G-OnRamp Education Article is published on PLOS Computational Biology
The G-OnRamp Application Note is published on Bioinformatics
See the preprint at bioRxiv
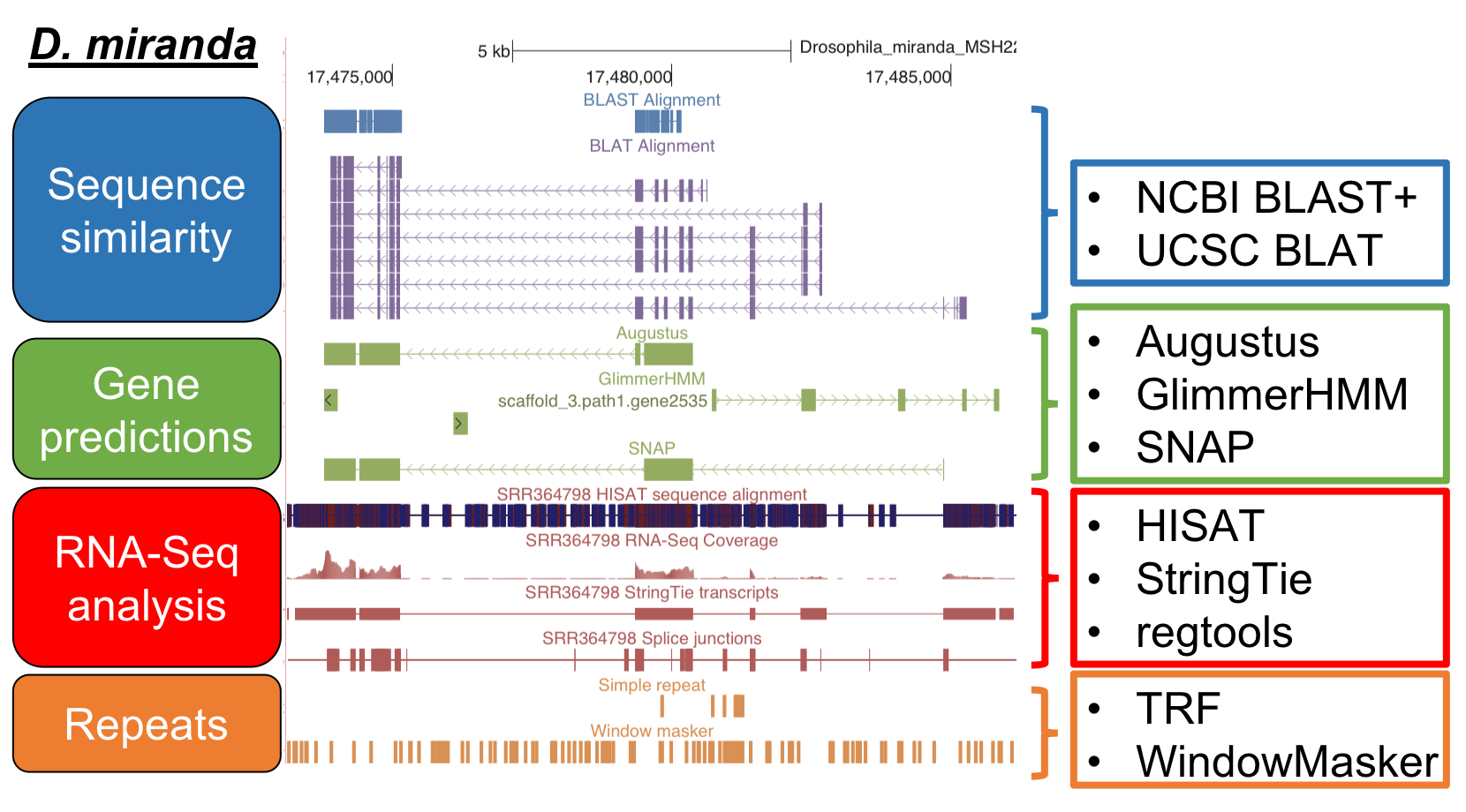
G-OnRamp tools
Sequence similarity
- NCBI BLAST+
- UCSC BLAT
Gene predictions
- Augustus
- GlimmerHMM
- SNAP
RNA-seq
- HISAT
- StringTie
- regtools
Repeats identification
- Tandem Repeats Finder
- WindowMasker
Apollo interaction tools
- Create or Update Organism
- Apollo User Manager
- Delete an Apollo record
Create Genome Browser Assembly Hubs
- Hub Archive Creator
- JBrowse Archive Creator